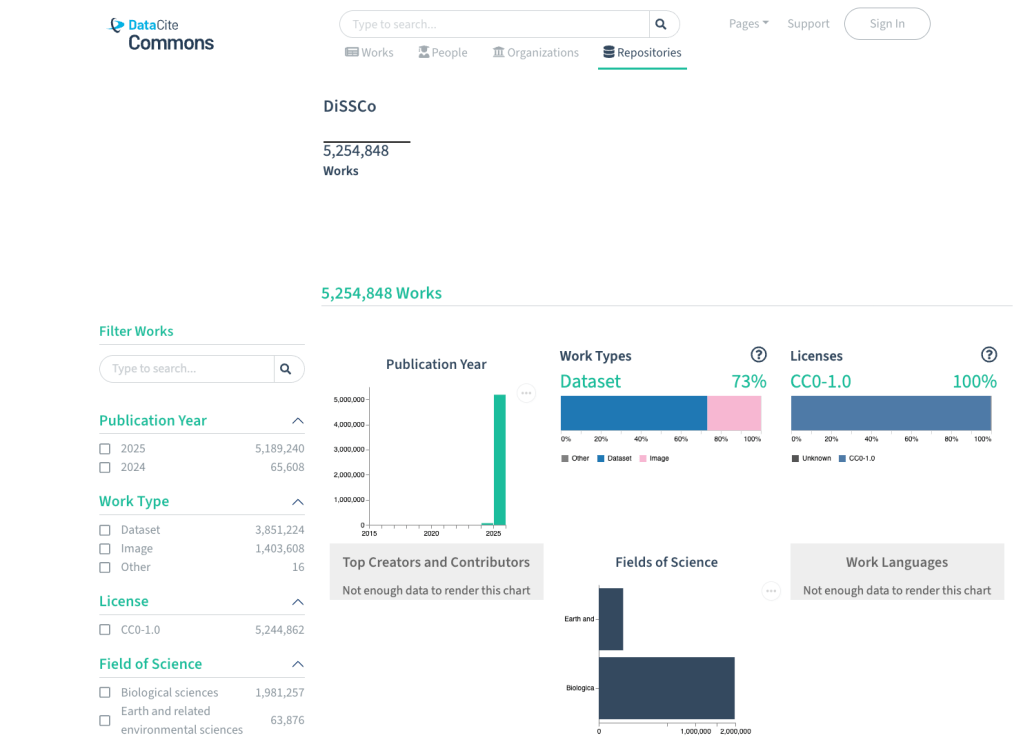
DiSSCo demos are back! It has been a while, and the team has gone through some changes, but that didn’t mean that development has halted. In fact, DiSSCo has hit an impressive milestone of over 5 million DOI’s minted. In additional to filling our production environment, we have some new and existing features to present.
DiSSCo Transition Project has officially ended, and one of the goals of DiSSCo was to deploy a first production version of the Digital Specimen Architecture (DSArch). With the release of a DiSSCo Minimal Viable Product (MVP), we are now able to start minting DOI’s for specimen and digital media. We are incredibly excited to share with you that we have recently hit the five million mark. For more information, please see our previous blog post.
Besides the DOI’s, we went over a number of other topics. Melanie de Leeuw gave us a presentation on the work she and Mathilde Lescanne are doing with the Naturalis user group. Their first focus is on improving the taxonomic annotation workflow. Together with Naturalis collection managers and taxonomic experts, they are testing and improving how experts can add new or modify existing taxonomic identifications through our DiSSCover interface. This is an excellent first step in onboarding our first experts, and we hope to inform you on the progress in our next session.
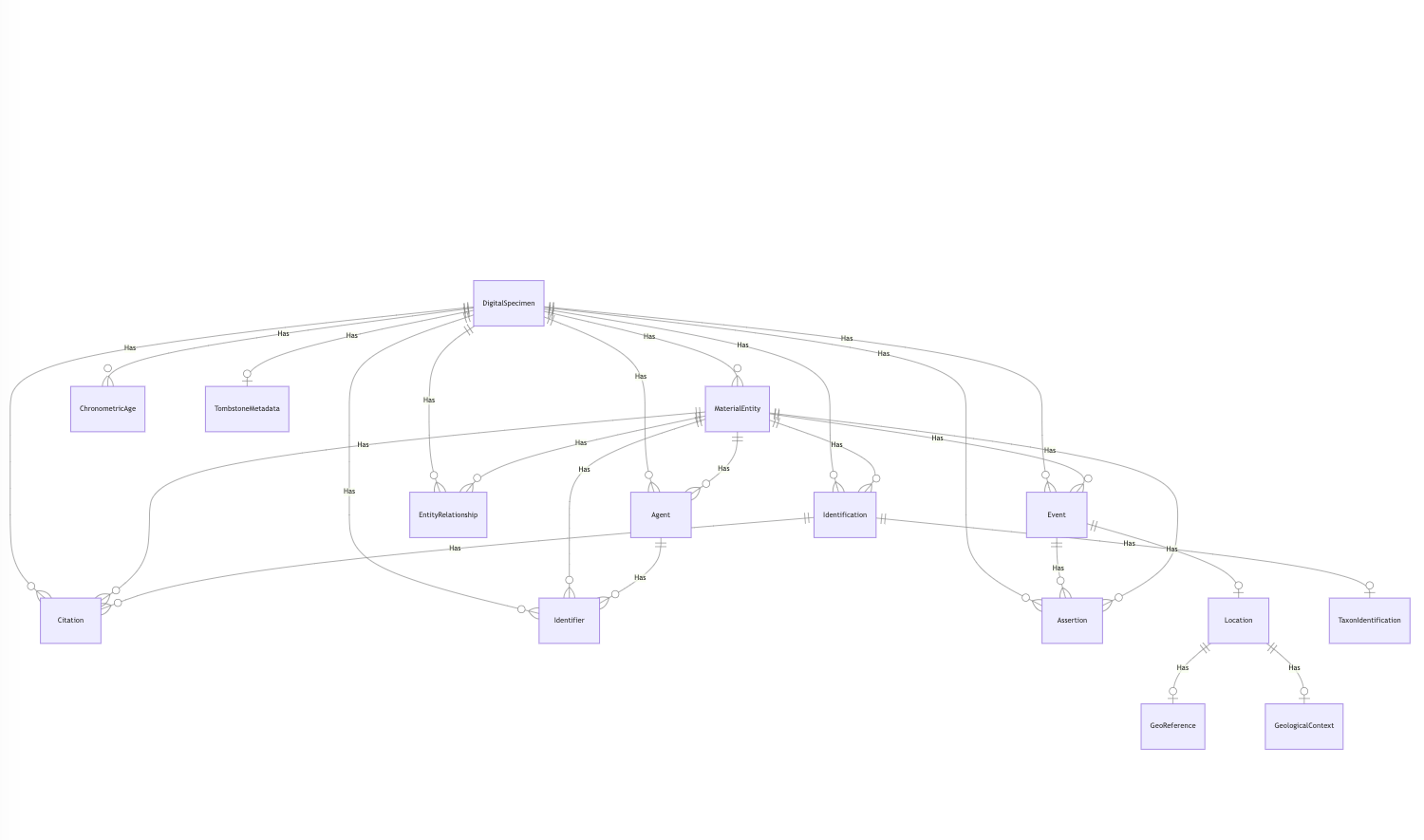
The next topic involved DiSSCo’s activity as early adaptor of the new Darwin Core Conceptual model and the associated Darwin Core Data Package (DwC-DP). DiSSCo has provided input for this new data standard and is at the forefront of implementation. Any data in DiSSCo will be available for download in the new DwC-DP. In addition, we also implemented an option to download any dataset in the current standard, the Darwin Core Archive (DwCA). Please be aware that the Darwin Core Conceptual Model will soon be in open review.
As the last topic, we wanted to show a new integration with a Machine Annotation Service (MAS). After community consultation we recognised VoucherVision, developed by William Weaver, as one of the leading tools for label transcription. As label transcription is an often manual time-consuming task vital to the digitisation of the specimen, we wanted to experiment if VoucherVision could help us. By integrating VoucherVision as a MAS in DiSSCo we are now able to run it on any specimen in our acceptance infrastructure.
We concluded the demonstrations with 15 minutes of questions. We would like to thank all participants and hope to see you at our next demo!
The following topics were presented in the demo:
- DiSSCo at past 5 million DOI’s
- User group starting on Taxonomic annotations through DiSSCover
- DwCA and DwC-DP export products from DiSSCo
- Voucher Vision integration in DiSSCo Sandbox
Looking forward to our next demo, which will be held in December, we hope to show the following topics:
- Ongoing improvements to data ingestion
- Move to 20 million DOIs
- Further improvements to annotation flow in DiSSCover
- Start testing with first experts
- MAS scheduling on ingestion
- (public) Virtual Collections (TETTRIs deliverable)
- DWCA and DwC-DP available as export in DiSSCover
- Refine annotation evaluation flow
- Image (derivatives) storage in DiSSCo
- Support integration with ELViS
- Switch underlying Identity and Access Management Infrastructure
Slides from the presentation can be downloaded here: https://docs.google.com/presentation/d/1bFm7iM1NJ72bomsx6FJJ48q6IGt_ZXhGjG1Ca9GvQIA/edit?usp=sharing