In biodiversity and environmental sciences, access to trusted, persistent, and citable data is key to accelerating research and discovery. That’s where Digital Specimen DOIs come in—a foundational tool for connecting your research to verifiable specimen data. Darwin Core supports them through a new term called “digitalSpecimenID” and Pensoft supports them in their ARPHA journal system. But what exactly are they—and how should you use them?
What is a Digital Specimen DOI?
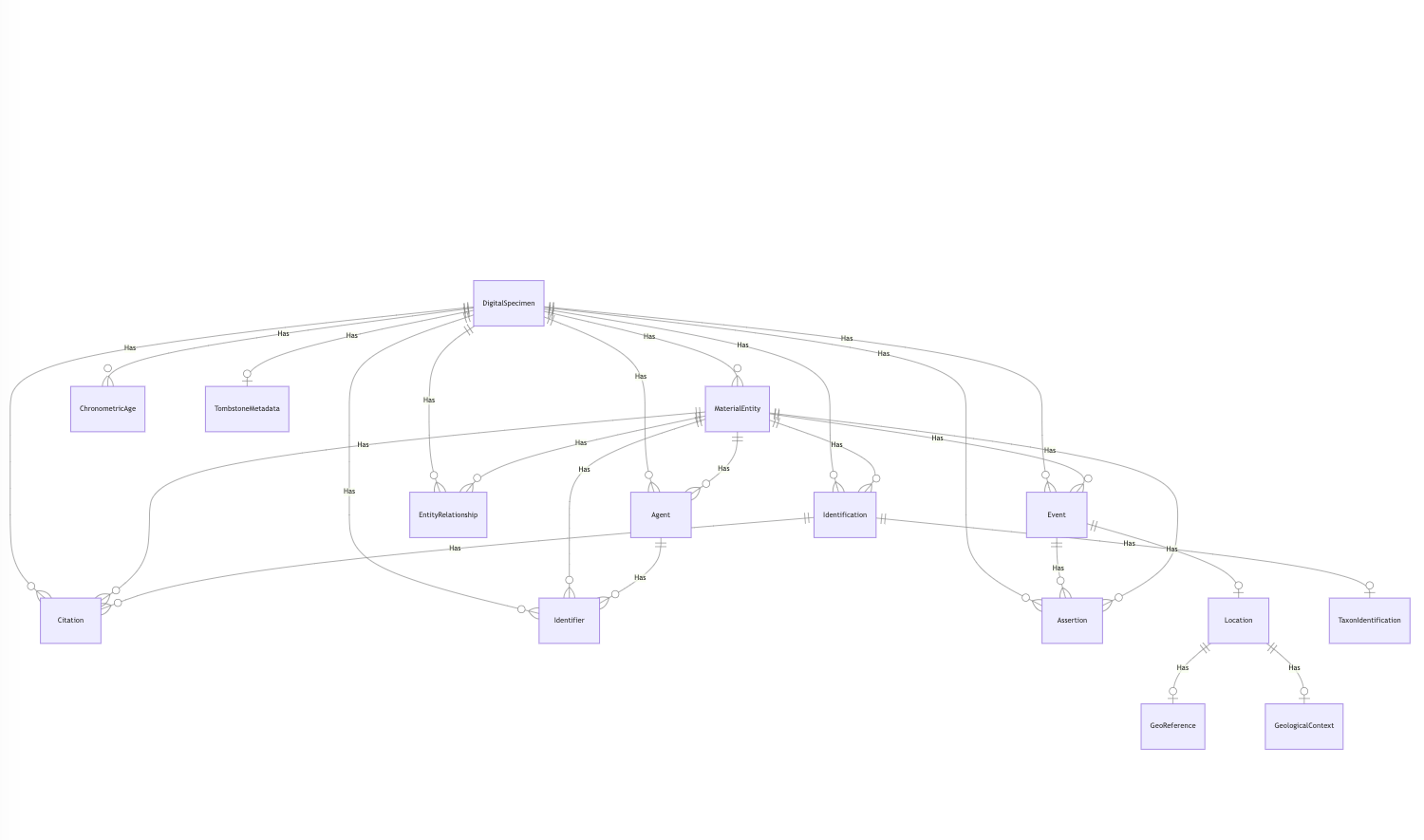
A Digital Specimen is versioned dataset providing a digital representation of a physical specimen (like a herbarium sheet, insect, or fossil) held in a natural science collection. It can be annotated, curated and extended with new data. A DOI (Digital Object Identifier) is a globally unique, persistent identifier that allows that specimen to be reliably referenced, cited, and accessed online.
Think of it as a permanent web address that always points to a specific digital specimen record—no matter where it’s hosted or how systems evolve. Digital Specimen DOIs allow you to:
🔗 Unambiguously reference specimens in publications and datasets.
🧬 Link your research outputs to the exact specimens used in your work.
📊 Track provenance, usage, and impact of specimen-based data over time.
Why Use DOIs for Specimens?
✅ Citable – You can cite specimens like research papers, improving credit for collectors, curators, and institutions.
🔍 Findable – DOIs are indexed by metadata aggregators such as DataCite and CrossRef, making your data easier to discover.
🔄 Interoperable – They help link specimen records to DNA sequences, publications, images, annotations, and more.
🛠️ Machine-Actionable – DOIs support automation and integration in digital workflows, critical for large-scale biodiversity research.
🔒 Persistent – Even if websites change, DOIs always point to the right data, helping ensure long-term accessibility.
Why Researchers Should Care
🧾 Cite specimens directly – Credit the physical evidence behind your research.
🔗 Integrate with nanopublications – Reference fine-grained data assertions (e.g., “Specimen X was observed with Trait Y in Location Z”) with full provenance.
📡 Enable reproducibility – Make your data and methods traceable and verifiable.
🧠 Power knowledge graphs – Link specimens to genes, traits, publications, and ecological observations.
🔄 Reuse with confidence – Know exactly where data came from and how it’s been used.
The advantages of DOIs
🔍 Findable – DOIs are indexed by metadata aggregators such as DataCite and CrossRef, making your data easier to discover and making your data connected in scholarly knowledge graphs.
🛠️ Machine-Actionable – DOIs support automation and integration in digital workflows, critical for large-scale biodiversity research. Digital Specimen DOIs are designed to always provide metadata and a machine readable version of the data (JSON) in addition to a human readable HTML page.
🔒 Persistent – Even if websites change, DOIs always point to the right data, helping ensure long-term accessibility. It is the mission of the DOI foundation and its registration agencies to ensure the resolvability of the DOIs.
How Do They Work?
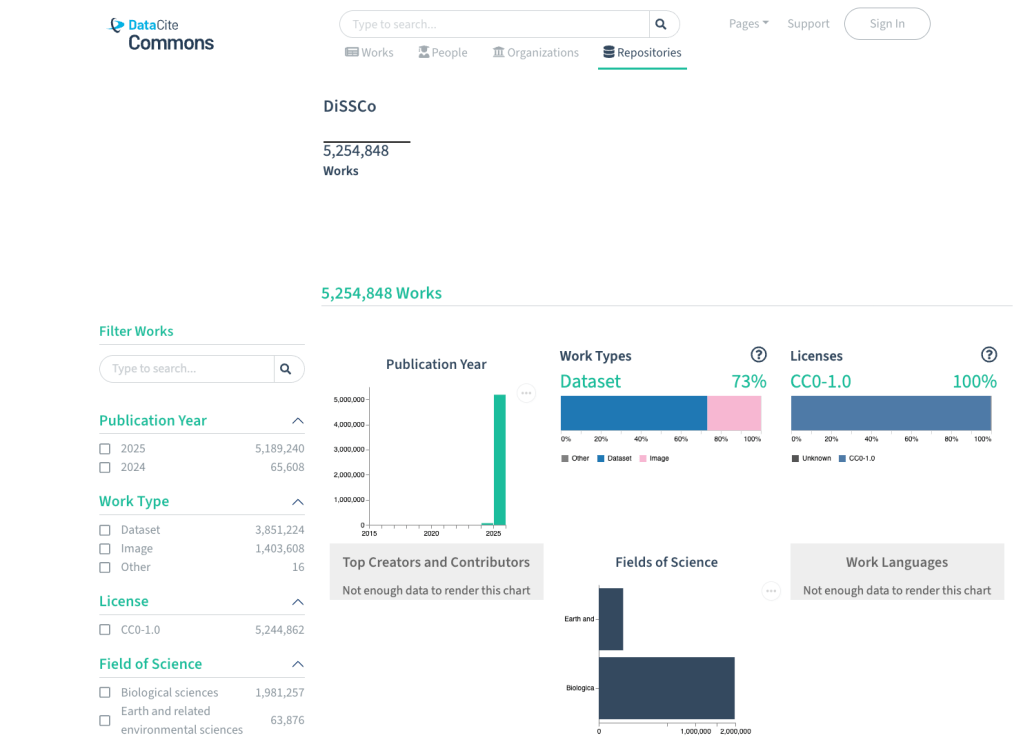
Behind the scenes, a DOI resolves via the Handle System, it links to a Handle record that automatically redirects to a HTML landing page for the specimen. The link to that landing page is included in the Handle record, so it can be updated when the URL for that landing page changes. DataCite provides DiSSCo with a prefix (currently only 10.3535) which DiSSCo uses to issue DOIs when digital specimens are published, and DiSSCo provides DataCite with the DOI metadata.
But a Digital Specimen DOI is designed to do much more than that, making use of advanced capabilities of the Handle system. it allows websites to provide metadata like the physical specimen catalog number as context to a user, with a tooltip as demonstrated here. It allows a user to directly go to a specimen catalog record if it exists online, using a locatt suffix: https://doi.org/10.3535/EF9-THJ-D7S?locatt=view:catalog. The same can be used to go directly to a JSON representation: https://doi.org/10.3535/EF9-THJ-D7S?locatt=view:json. Pretty cool. And while the DOI by default redirects to the latest version of the digital specimen, it is also possible to refer to a specific version by appending the version number, for example for version 2: https://doi.org/10.3535/EF9-THJ-D7S?urlappend=/2.
Ready to Get Started?
If you’re managing specimens or working with specimen data, consider using the digital specimen DOIs. It’s a small step with a big impact—for you, your data, and the global research community.
👉 Tip: Include digital specimen DOIs in your methods sections, data citations, and supplementary materials for greater impact and reproducibility. For an example, see this publication: https://doi.org/10.3897/BDJ.12.e129438.
Always include DOIs (and any other persistent identifiers) with the full URL, so including “https://doi.org/”. If that is not possible, use “doi:” instead. Note that the specimen images also have a DOI that you can use. The digital specimen will give an example of how to cite it, for example:
🔗Naturalis Biodiversity Center (2025). Animalia. Distributed System of Scientific Collections. [Dataset]. https://doi.org/10.3535/5BP-R3Z-6K2.
If you publish your data in Darwin Core, include the DOI using the (new) term digitalSpecimenID. This allows aggregators like GBIF to link to the digital specimen.
Check if your data sources provide DOIs for digital specimens. If not, encourage data providers to adopt them—many already integrate DOI assignment as part of digitization workflows.
Nanopublications: Going One Step Further
For researchers working at the frontier of semantic data publishing, nanopublications offer a way to formally express and cite individual assertions—like taxonomic identifications, ecological observations, or annotations on a specimen.
When combined with Digital Specimen DOIs, nanopublications allow you to:
- Reference atomic facts with full provenance.
- Enable machine-readable assertions.
- Support automated reasoning across data networks.
Learn more at nanopub.org and explore how to publish assertions connected to digital specimens or look at https://doi.org/10.3897/BDJ.12.e129438 for an example. Let’s take the Assertion: “Specimen RMNH.ARA.18251 is Phintelloides scandens sp. nov.”
A Nanopub enhances the semantic precision of this assertion by including author credit and provenance metadata, and provides a machine-readable format enabling automated reasoning.
Persistent data builds lasting knowledge. Start leveraging Digital Specimen DOIs today—your future self (and your citations) will thank you.