Dear Readers,
In this post, we would like to update you on some of the recent technical advancements that we have made. Our team has been working tirelessly on various aspects including PID infrastructure development, data modeling work, and both frontend and backend development of the DiSSCover interface. These updates have allowed us to enhance the functionality and overall user experience. These are significant steps toward realising the vision of DiSSCo.
DiSSCover Sandbox interface: Our DiSSCo sandbox has gone through several iterations (both front and backend). We added different search, filter and compare features to easily find the Digital Specimens: 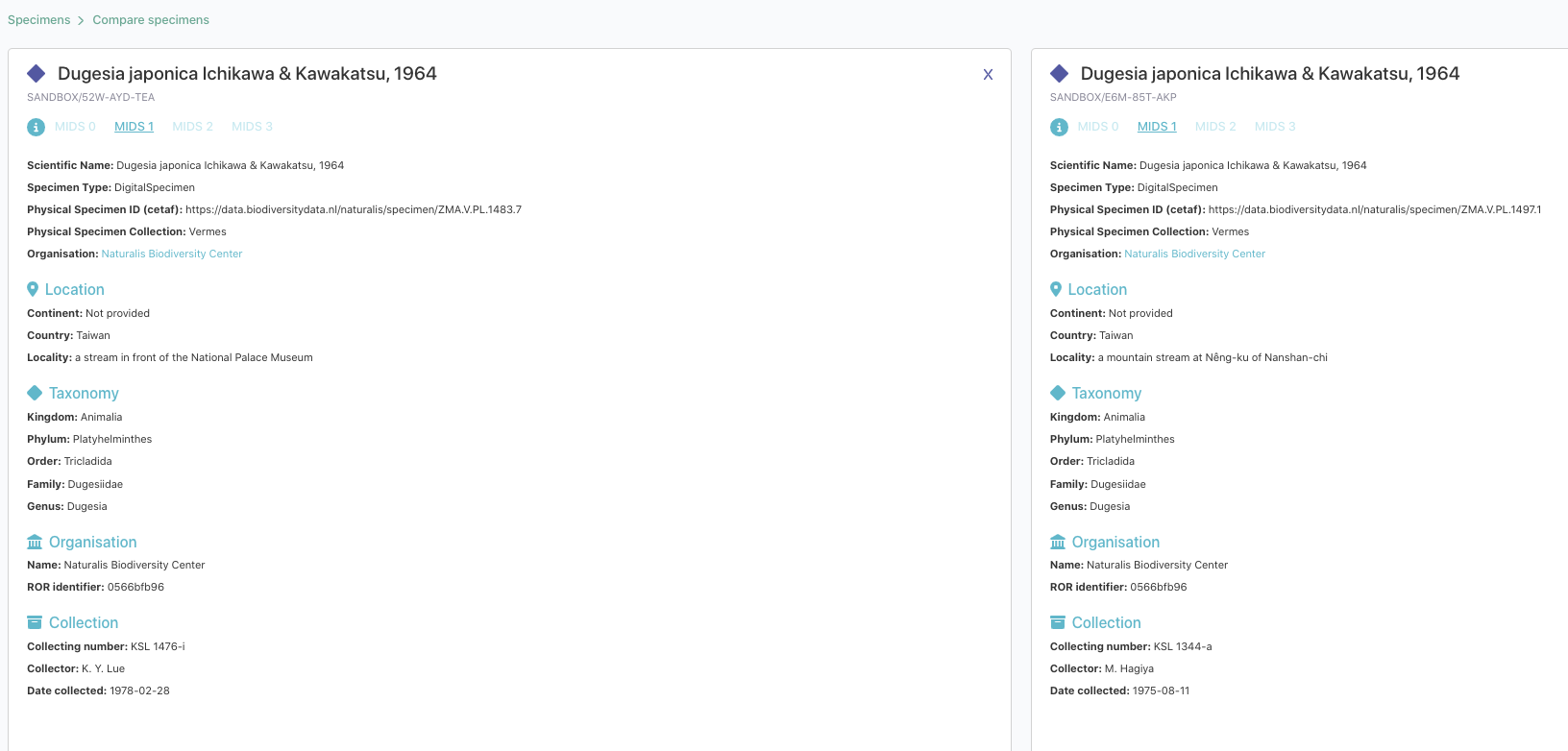
We also created a user onboarding tool to guide you through these features. Please let us know and use Github to send feedback.
Authentication: With the help of our partner at GRNET, we have incorporated ORCID and institutional login (with Keycloak). We are now working on the user onboarding workflow, login policy, and authorisation matrix.
Annotation: After authentication, the user can annotate either a full Digital Specimen record or a specific field. We have done work on creating a new Annotation data model (see the latest version of the Annotation JSON schema ). An RFC has been sent out to the community for feedback. We have made an effort to incorporate the W3C Web Annotation data model to use concepts such as Target, body, and motivation. Our current implementation allows for creating annotations based on this schema and these annotations are created as FAIR Digital Objects (with PID and FDO Records). We are working on adding a batch feature and also a scoring and trust model. 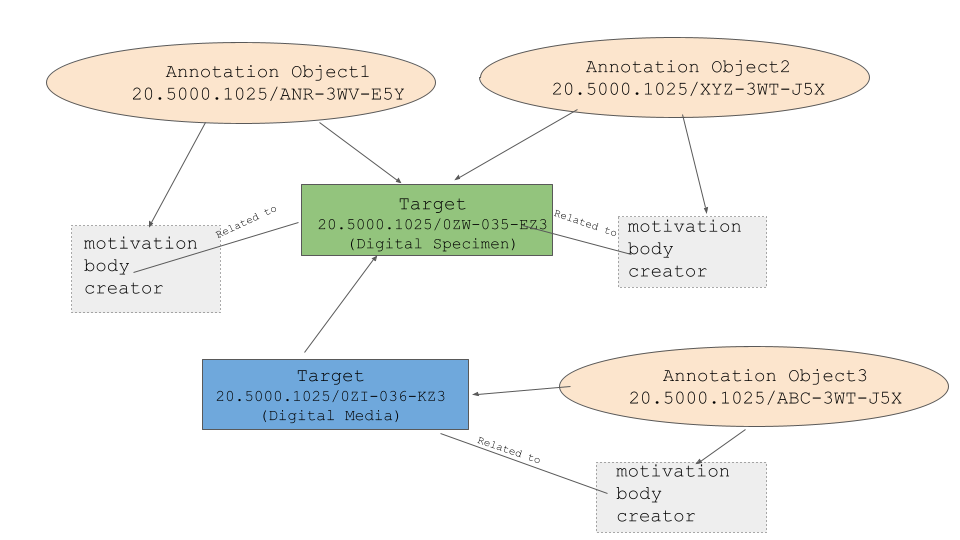
Data Harmonisation and GBIF Unified Data Model: As we have different data standards, formats, and terms, they introduce issues in the data lifecycle. This has an impact on data quality and building trust between data providers and consumers. So from the beginning, we are aware of the pitfalls and have made significant efforts in term/vocabulary harmonisation by reusing existing namespaces and ontologies as much as possible. We are also working closely with the GBIF Unified Data Model and the TDWG MIDS specification to incorporate these elements into openDS. When a new Digital Specimen is created, this approach can help us connect and move objects between different platforms easily. For example, the GBIF data model has an “EntityRelationship” component that provides a way to make connections between different entities. Instead of creating our own entity relationship model, we have incorporated it within the Digital Specimen data structure. Here is a snippet of our JSON serialisation that shows openDS attributes (ods), combined with Darwin Core (dwc) and also elements from the GBIF data model. Even though in this example, our initial sandbox implementation is duplicating some items (such as the ROR for the institutionId), the idea is that we now have a structure to build upon and add relevant, newly discovered links for the Digital Specimen thus making it a dynamic, actionable object.
"attributes": {
"digitalSpecimen": {
"ods:id": "https://doi.org/TEST/WY0-JT7-5JG",
"ods:version": 1,
"ods:created": "2023-10-16T11:46:26.577829Z",
"ods:midsLevel": 1,
"ods:normalisedPhysicalSpecimenId": "https://herbarium.bgbm.org/object/BW02635010",
"ods:physicalSpecimenId": "https://herbarium.bgbm.org/object/BW02635010",
"ods:physicalSpecimenIdType": "Resolvable",
"ods:topicOrigin": "Natural",
"ods:topicDomain": "Life",
"ods:topicDiscipline": "Botany",
"ods:hasMedia": true,
"ods:specimenName": "Spermacoce scabra Willd.",
"ods:sourceSystem": "https://hdl.handle.net/TEST/HVJ-A25-XYN",
"ods:livingOrPreserved": "Preserved",
"dcterms:license": "http://creativecommons.org/publicdomain/zero/1.0/",
"dcterms:modified": "1465900920000",
"dwc:basisOfRecord": "PreservedSpecimen",
"dwc:preparations": "herbarium sheet",
"dwc:institutionId": "https://ror.org/00bv4cx53",
[..]
entityRelationships": [
{
"entityRelationshipType": "hasOrganisationId",
"objectEntityIri": "https://ror.org/00bv4cx53"
},
{
"entityRelationshipType": "hasSourceSystemId",
"objectEntityIri": "https://hdl.handle.net/TEST/HVJ-A25-XYN"
},
{
"entityRelationshipType": "hasFdoType",
"objectEntityIri": "https://doi.org/21.T11148/894b1e6cad57e921764e"
},
{
"entityRelationshipType": "hasPhysicalIdentifier",
"objectEntityIri": "https://herbarium.bgbm.org/object/BW02635010"
},
{
"entityRelationshipType": "hasLicense",
"objectEntityIri": "http://creativecommons.org/publicdomain/zero/1.0/"
}IIIF Support and Image Annotation: As IIIF is a well-established community standard for image interoperability, we are working on support for that in our sandbox. There are different types of media objects linked to the specimen. We are also working image annotation feature which will align with the current annotation data model.
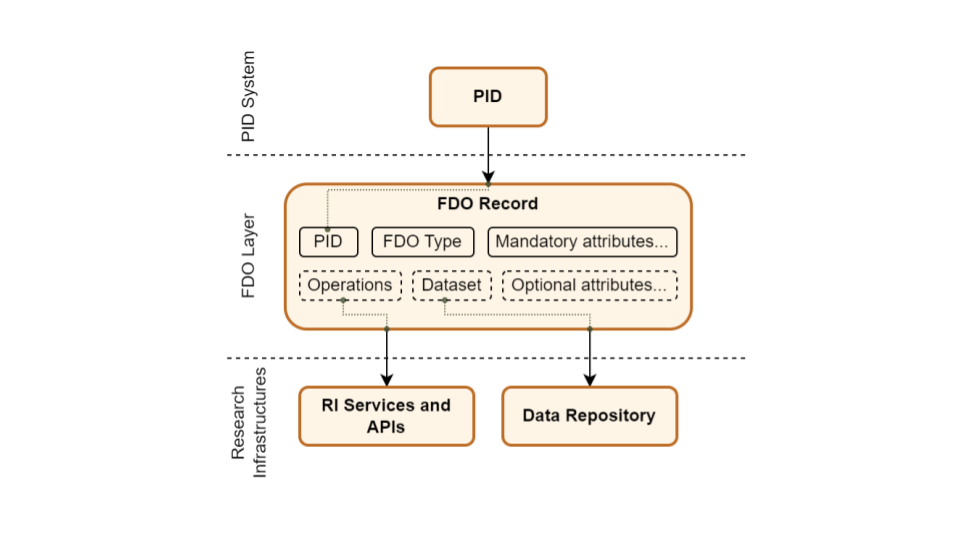
Persistent Identifiers and DOI infrastructure: A lot is going on in this area, as PID is one of the core components for DiSSCo infrastructure and FAIR implementation. Please see our other blog post that talks about the test PID infrastructure for more technical details.
Biodiversity Digital Twin Project and FAIR Digital Objects: One of the design principle we are trying to adhere is to re-use existing components and also thinking about cross project and cross research infrastructure alignment. This way we avoid duplication of efforts. The FAIR Digital Object (FDO) work within the BioDT project is a great example of this. We are now working on creating FDO Profiles for different Digital Objects that will be part of BioDT (similar to the DiSSCo FDO Profile). This is to understand how FDO can fit into the BioDT project as an abstraction layer between different infrastructures. The main benefit of this approach is achieving interoperability as different datasets and models will be needed to create digital twins.
Conference presentations, hackathon, and publications
A DiSSCo poster was featured at the First Conference on Research Data Infrastructure (CoRDI). At the conference, we presented some of the lessons learned during the design and preparatory phases of DiSSCo. You can view the poster online on the Zenodo platform.

At the TDWG 2023 conference, we delivered several presentations covering a range of topics, including data linking and persistent identifiers, a recipe for integration AI services into the DiSSCo infrastructure, mapping and MIDS, and data harmonisation.
We are actively engaged in the global discussion about the implications of the FAIR Digital Object paradigm for biodiversity research. A new paper proposes leveraging the FAIR Digital Object framework to advance the global vision of the Digital Extended Specimen concept. This paper emerged from discussions that took place at the 2022 FAIR Digital Object Forum conference. We are eagerly anticipating the 2024 FDO conference in Berlin. We are also excited about the upcoming 2024 FDO conference in Berlin.
This week, we are participating in the Elixir biohackathon, where we will be exploring improved linking from sequence data to specimen and sample repositories.
Keep an eye on our GitHub page and stay tuned for more updates.
